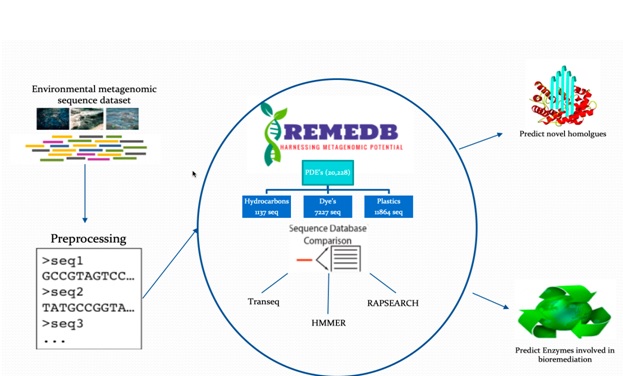
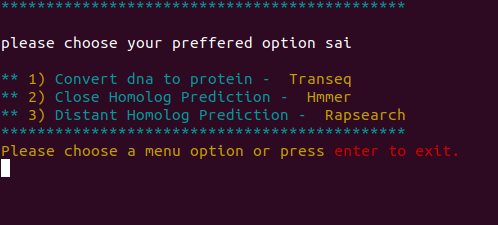
Welcome to RemeDB
RemeDB aims at identifying Pollutant Degrading Enzymes (PDE) from metagenomic sequences.
Enables classification and prediction novel enzymes classes.
User interprtable outputs generated at lightning fast speed ( number of computer threads )
Enzymes associated with bioremediation were categorized under 3 categories:
> Hydrocarbon degrading enzymes
> Plastic degrading enzymes
> Dye degrading enzymes